Oceanographers love MATLAB\(^\circledR\) for processing oceanographic data. I have no explanation as to why they love Matlab. I suppose it’s been the de facto standard within the field for a long time, and legacy is a big deal. I have used Matlab for data processing and plotting for several years. With MatLab I implemented a vast amount of tools and rather completed numerical models. Being a self sponsored graduate student at an institution that doesn’t have a subscription, I found sticking to Matlab not only is expensive, but also unsustainable. A chunk of my scripts just sleeping in my drive because I can no longer run the software in my machine.
The Matlab was the core software for my PhD until recent, when I switched to R because of the license issue. The other reason is R as some cool packages that are far easier to use for exploratory and producing publication-quality graphics. Though, I found myself frustrated when learning R because I was doing Matlab syntax in R—which lacked the flexibility of MATLAB. When I learned to unlearn Matlab and learn R syntax, it just took me some weeks to stand up and walk.
In this article, I’m going to illustrate how to process oceanographic data with oce package in R. oce package has some cool features for reading, processing and plotting oceanographic data. It is written by Dan Kelley and Clark Richards (2018). oce support many oceanographic data but for this post I will only focus on the CTD measurements. I need oce and ocedata (Kelley, 2015) package to process the CTD data in R (R Core Team, 2018).
require(oce)
require(ocedata)On February, 2018, The Tanzania Fisheries Research Institute (TAFIRI) had a field campaign to collect CTD data using a Sea-Bird SBE19plus equipment in Rufiji River. I voluntered for the field campaign and got some CTD profile data that I use to show you how to process and manipulate CTD data and plotting the profiles and section with oce package in R.
Importing CTD Data
SBE comes with SBE Data Processing Graphic User Interface to help converting the data from binary to engineering unit. I use this software to convert .hex raw data from CTD and store them in .cnv file format. Once the file was ready, I imported it from local folder into R workspace with read.ctd() function. according to Kelley & Richards (2018)
read.ctd()is a base function that read different format of CTD data includingread.ctd.odffor the ODF data used in Fisheries and Oceans (Canada),read.ctd.wocefor data in World Ocean Circulation Experiment format,read.ctd.woce.otherfor a variant of WOCE data,read.ctd.itpfor ice-tethered-profiler data, orread.ctd.sbefor Seabird data.
#read profile of station1
stn1 = read.ctd("./ctd_rufiji/SBE19plus_01906740_2018_02_13_0885.cnv")oce organize the CTD object into three slots—metadata, data and processing log.
metadata: a list with information about the data like units, quality, sampling location, sampling time, …etcdata: a list of raw profile data like pressure, temperature, salinity.processinglog: a document track all the processing information
str(stn1,2)## Formal class 'ctd' [package "oce"] with 3 slots
## ..@ metadata :List of 27
## ..@ data :List of 13
## ..@ processingLog:List of 2Depending on configuration, the content in CTD file differs . But the data slots often remain fairly constant with the content. For instance, here we see the data slots with thirteen variables and six hundreds and sixteen observations for profiles in station_1.
stn1@data%>%as.data.frame()%>%dplyr::glimpse()Observations: 616
Variables: 13
$ pressure <dbl> 0.074, 0.074, 0.063, 0.074, 0.074, 0.074, 0.064, ...
$ temperature <dbl> 29.6868, 29.6889, 29.6911, 29.6922, 29.6925, 29.6...
$ conductivity <dbl> 0.008290, 0.008492, 0.008459, 0.008810, 0.008703,...
$ salinity <dbl> 0.0419, 0.0427, 0.0426, 0.0440, 0.0435, 0.0444, 0...
$ oxygen <dbl> 3.6746, 3.5385, 3.9256, 3.5252, 3.4002, 3.3936, 3...
$ fluorescence <dbl> 0.5136, 0.5091, 0.5068, 0.5144, -0.0227, -0.0166,...
$ bottlesFired <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ time <dbl> 0.00, 0.25, 0.50, 0.75, 1.00, 1.25, 1.50, 1.75, 2...
$ time2 <dbl> 33.72039, 33.72040, 33.72040, 33.72040, 33.72040,...
$ density <dbl> 995.7750, 995.7749, 995.7741, 995.7749, 995.7745,...
$ depth <dbl> 0.074, 0.074, 0.063, 0.074, 0.074, 0.074, 0.063, ...
$ descentRate <dbl> -2.632e-17, 0.000e+00, -3.000e-03, -2.000e-03, -1...
$ flag <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...Visualising Profiles
Looking on those number seems boring, let us take a quick gander at those number by making profile plots. Figure 1 shows the profiles of CTD measurements at station one. At top left and right we have some waer column profiles of temperature, salinity and density. At the bottom left is the temperature-salinity plot with contours of density.
#plot
stn1%>%plot()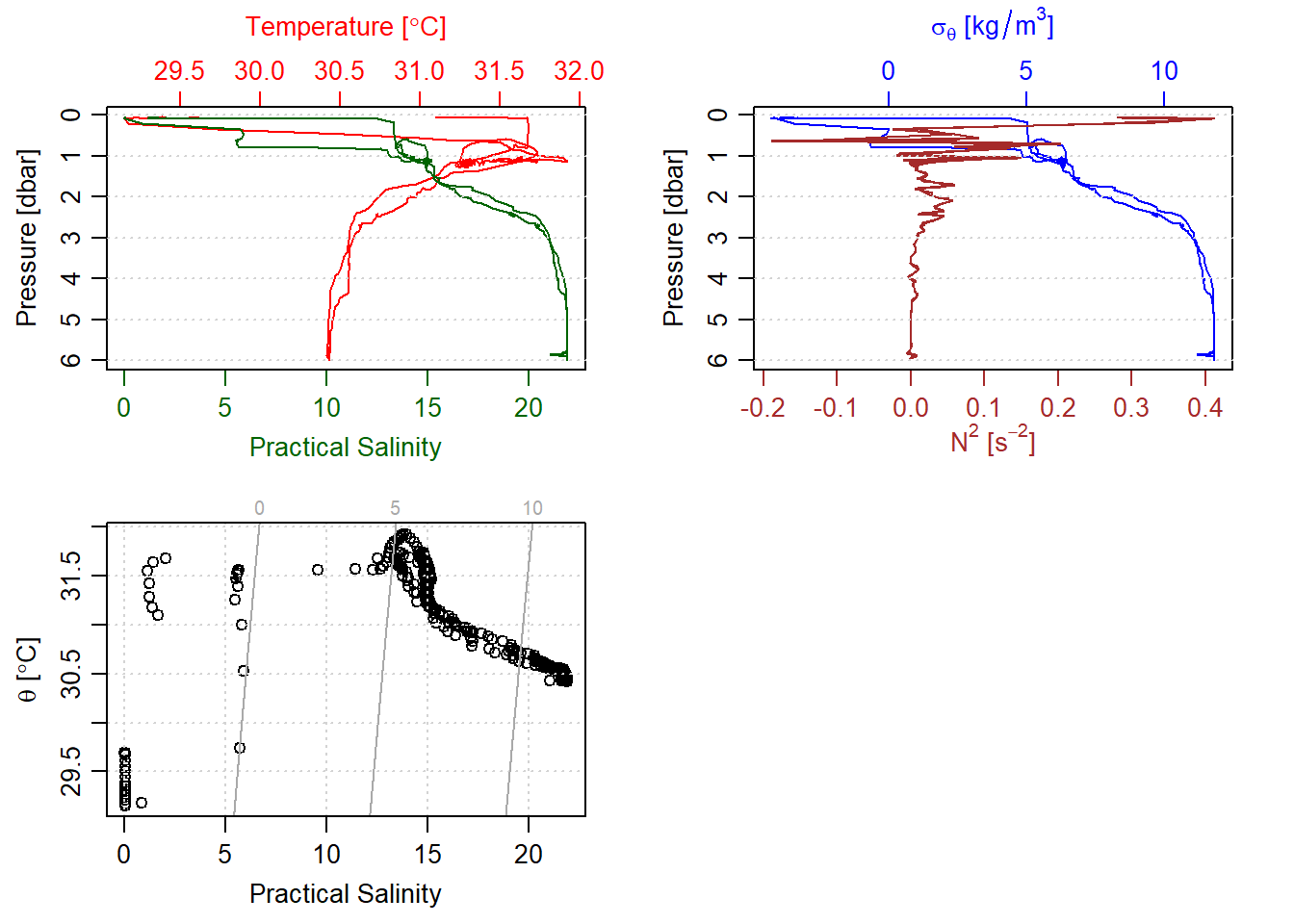
Figure 1: Unprocessed CTD measurement
Unfortunately CTD data for station one does not have a location data. This is because GPS information was not integrated into the SBE equipment. However, the location information were collected separately using a handheld GPS unit. I added this information manually for the station one.
cast.locations = read.csv("./ctd_rufiji/Rufiji_cast.csv")
stn1[["longitude"]] = cast.locations$Lon[1]
stn1[["latitude"]] = cast.locations$Lat[1]Plotting the CTD data that contain location information, often times plot a map showing the location of the CTD cast. Figure 2 is similar to to 1, except the bottom right has been added in this plot, which show where and when the cast was done.
stn1%>%plot()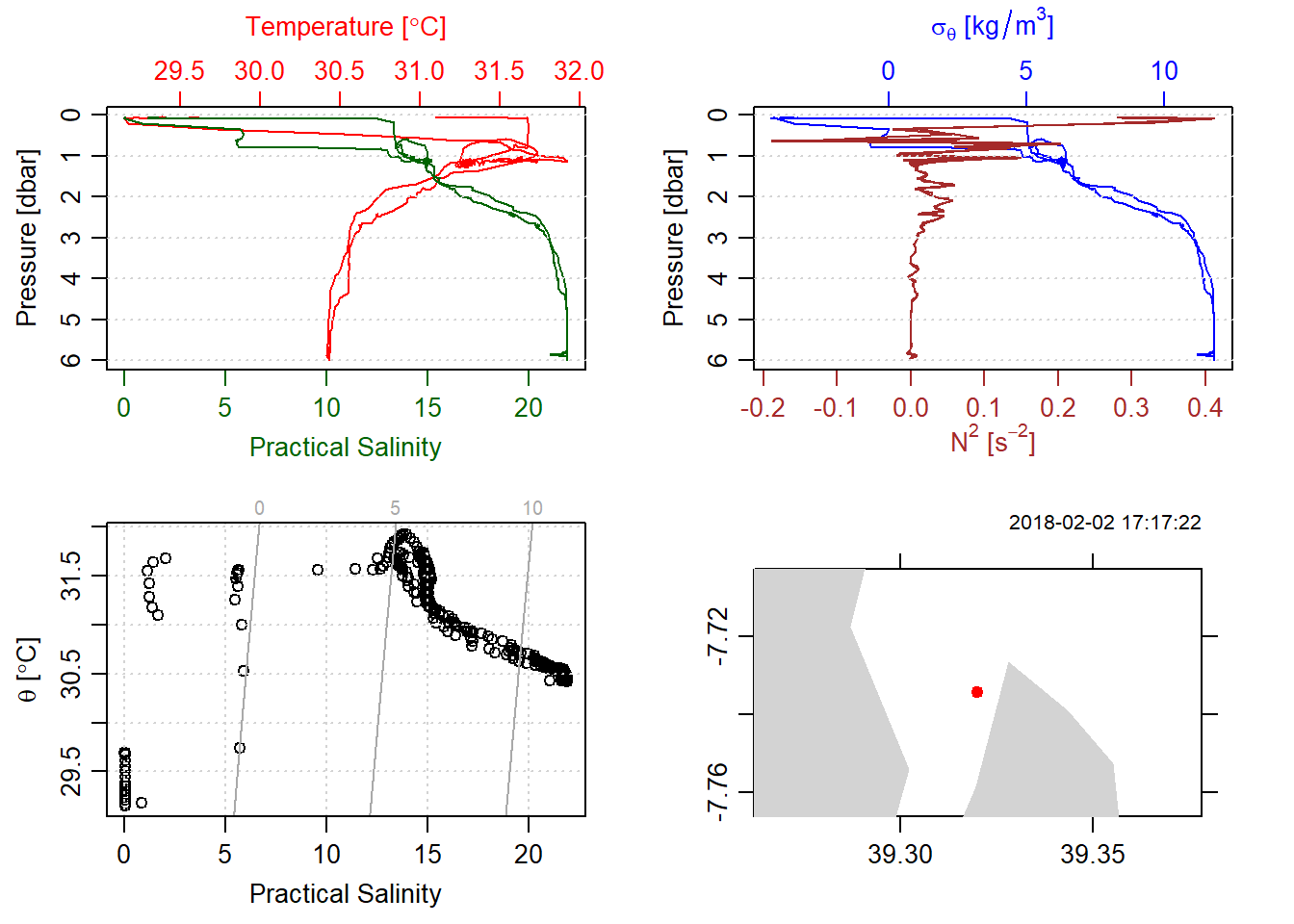
Figure 2: CTD data with location information
When we glimpse the salinity and temperature profile (Figure 3), we can see that there are profile measurements from downcast and upcast. Often in CTD profiling, the goal is to retain the downcast and drop all measurement from the upcast. The other thing you notice for figure 3 is the downcast and upcast profile of temperature and salinity are not smooth and sharp. This is because the rate of towing CTD instrument differs with because it was done manually.
par(mfrow = c(1,2))
stn1%>%plot(which = "salinity")
stn1%>%plot(which = "temperature")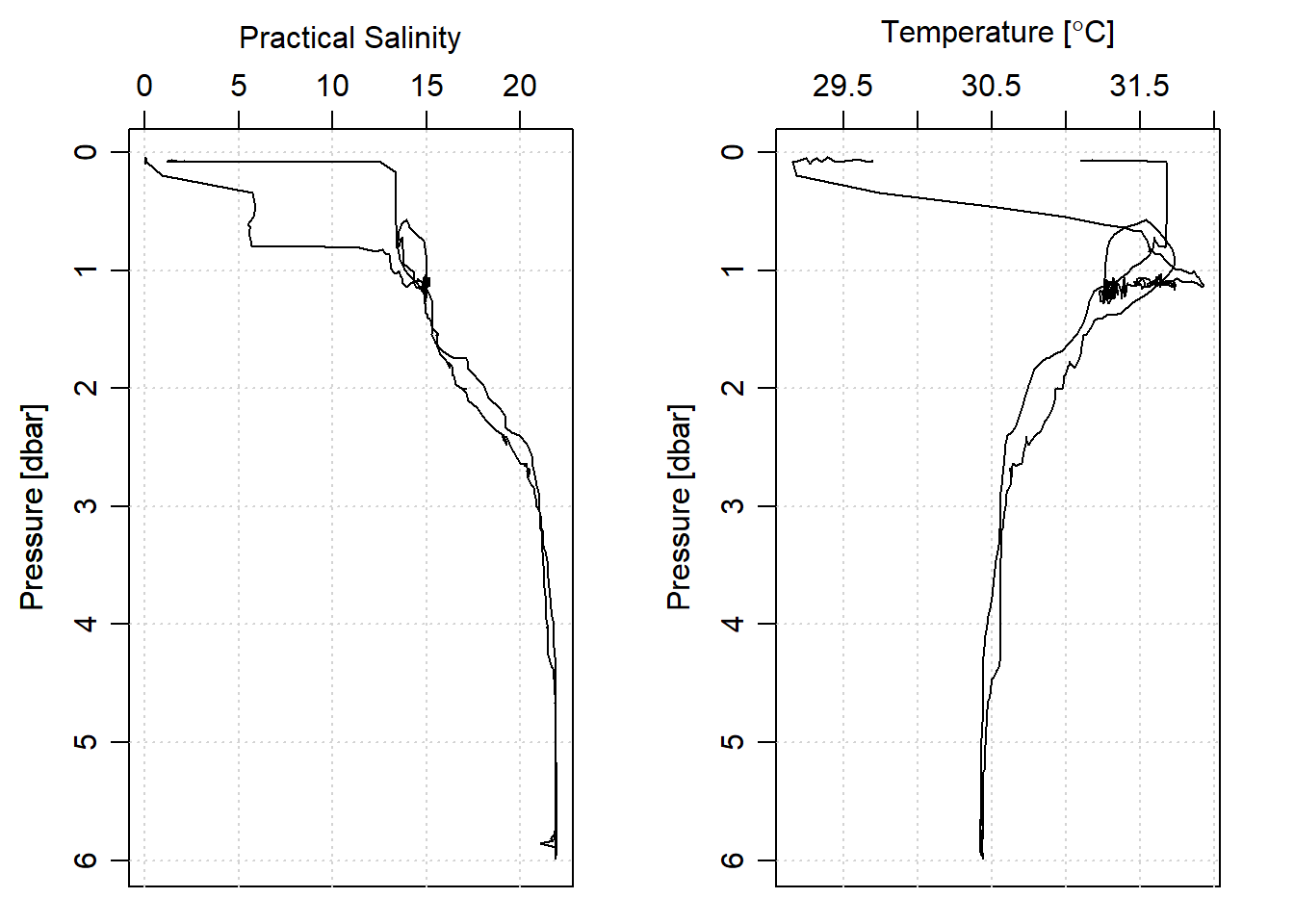
Figure 3: Salinity and temperature profiles showing downcast and upcast measurments
I removed the upcast CTD profiling with the ctdTrim() function and retained the downcast profiles. Once the upcast was dropped, I removed the artifacts introduced by towing rate. This was done by aligning the CTD measurements into the standard depth of 20 cm from the surface to the maximum depth of the cast with ctdDecimate() function. This produced elegant looking salinity and temperature profiles (Figure 4)
stn1.downcast = stn1%>%
ctdTrim(method = "downcast")%>%
ctdDecimate(p = 0.2)par(mfrow = c(1,2))
stn1.downcast%>%plot(which = c("salinity"))
stn1.downcast%>%plot(which = c("temperature"))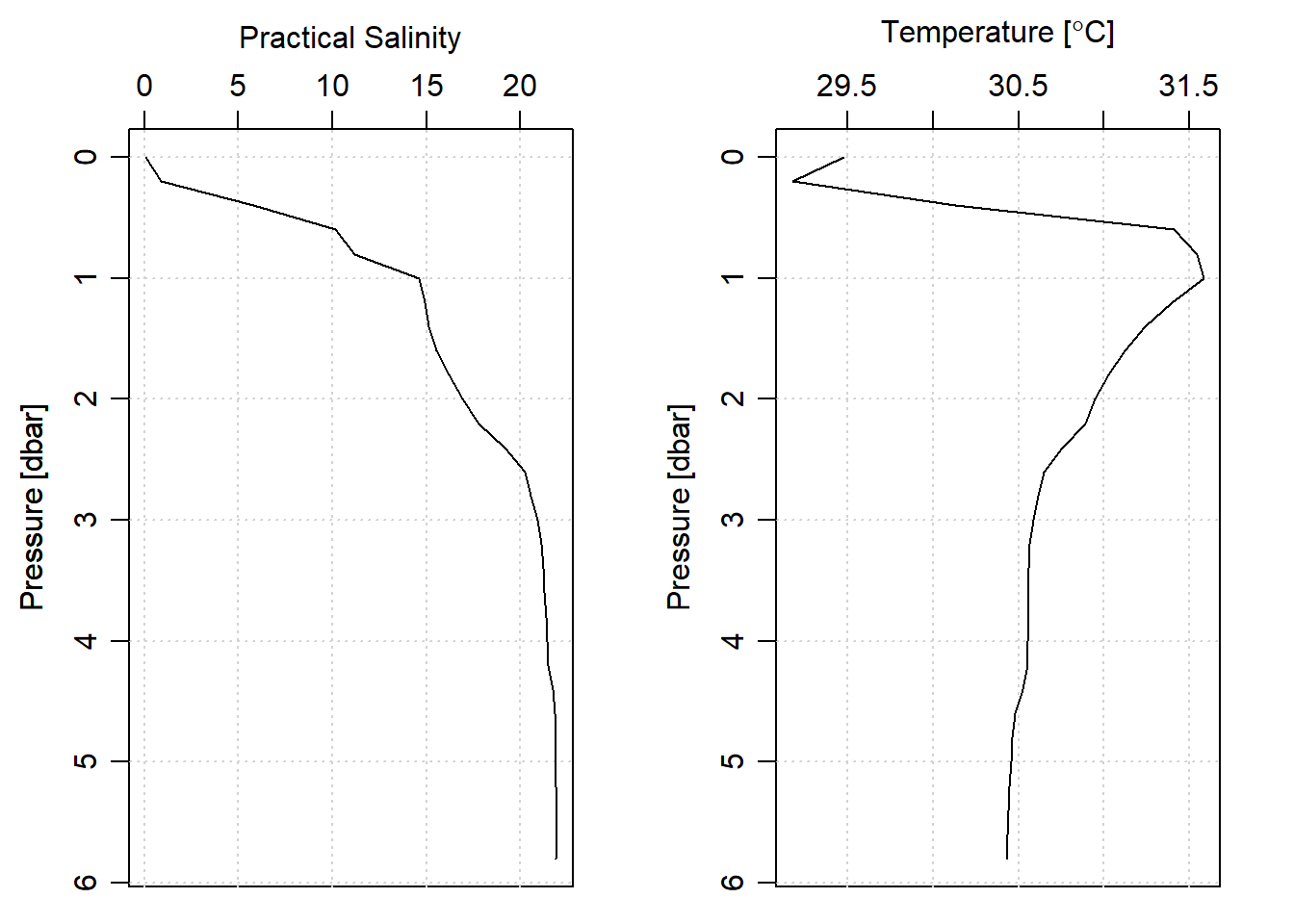
Figure 4: Profile o Salinity and temperature from downcast measurments and aligned to standard depth
plotting hydrographic section
The procedures above was just for one station, but hydrographic section involves multiple profiles. Therefore, we will loop through all the stations with a for() loop to read the `.cnv’ file, add the latitude and longitude for each cast
# create a list of cnv files
files = dir("./ctd_rufiji/", full.names = TRUE, pattern = ".cnv")Before looping, I preallocated a list files that will store each processed CTD cast in a list. The chunk below show the processed of looping CTD data. In a nutshell, it first read the file, then remove the upcast profile and align the profile measurement to a standard pressure of 20 centimeters. Then add the longitude and latitude information for each cast.
# loop through the files
ctd = list()
for (i in 1:18){
ctd[[i]] = read.ctd(files[i])%>%
ctdTrim(method = "downcast")%>%
ctdDecimate(p = 0.2)
ctd[[i]][["longitude"]] = cast.locations$Lon[i]
ctd[[i]][["latitude"]] = cast.locations$Lat[i]
ctd[[i]][["stations"]] = cast.locations$Name[i]
}Once we have a list of all the files, we can create a section along the river that only use the center positions of each cast. We notice that the CTD measurements was done at the middle of the within the channel with depth ranged from 3 to 9 meters.
section = list(ctd[[2]],ctd[[5]],ctd[[8]],
ctd[[12]],ctd[[14]],ctd[[18]])%>%
as.section()
sectionUnnamed section has 6 stations:
Index ID Lon Lat Levels Depth
1 39.316 -7.735 44 9
2 39.313 -7.757 43 8
3 39.305 -7.768 37 7
4 39.296 -7.781 32 6
5 39.287 -7.795 16 3
6 39.274 -7.809 20 4Figure 5 show the hydrographic section of temperature, salinity and oxygen within a distance of 10km from the mouth landward in the Rufiji River. Its pretty neat that we can see that the salinity decrease from the river mouth landward. This is because we did CTD casts during the high tide—when saline water was coming inland pushing freshwater further upland. We also see that the river has very low oxygen at just 2 meter from the surface that extend from the mouth to 10 km distance. The temperature shows uniform from the surface to the bottom along the section.
# par(mfrow = c(1,2))
section%>%plot(which = c("map", "temperature", "salinity", "oxygen"),
ztype = "image", showStations = TRUE)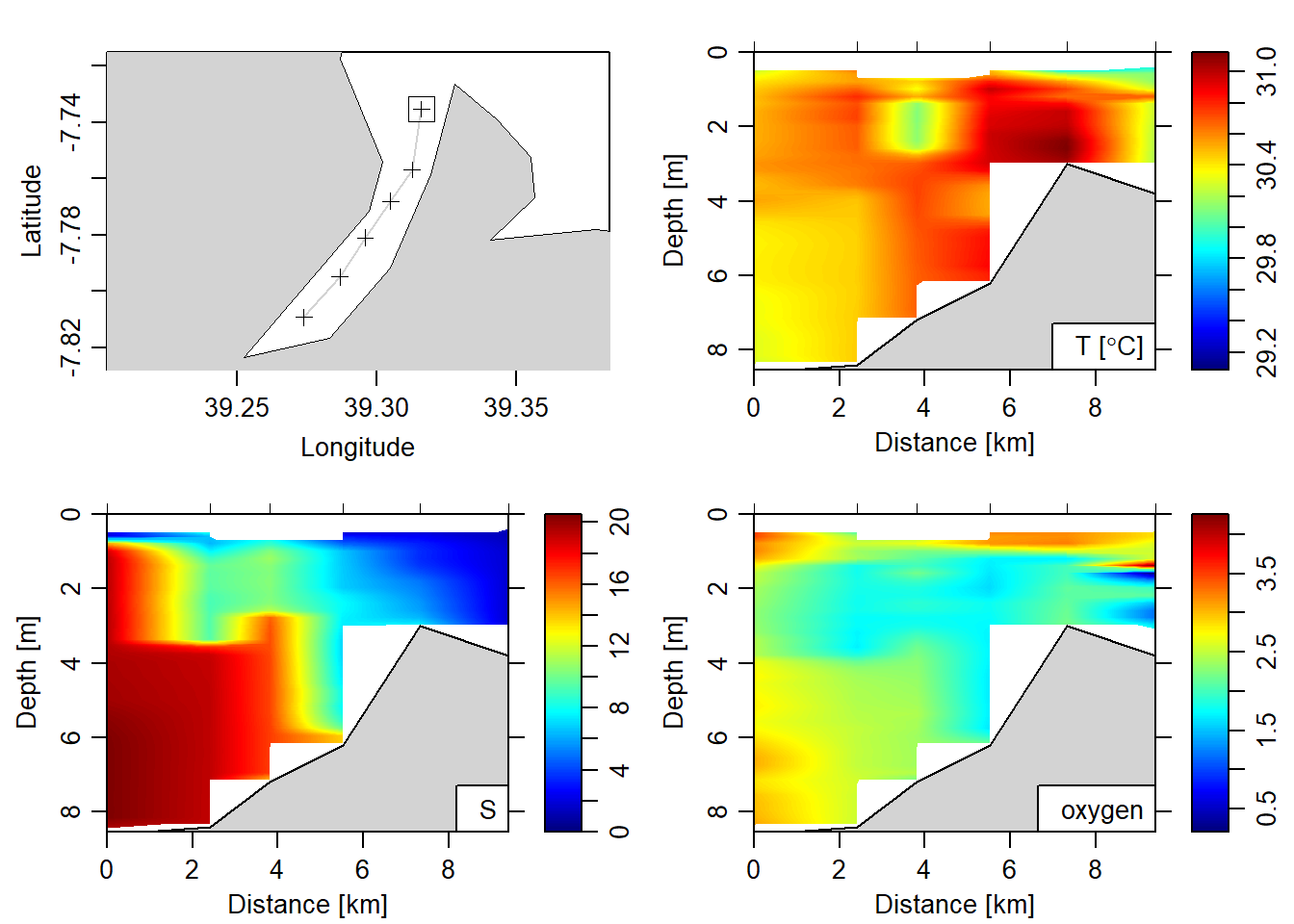
Figure 5: Hydrographic section of Rufiji River
Conclusion
In this post, we have seen the basics of processing CTD data in R with oce package. You have learnt how to import the data, manipulate and proting profiles and section.
Literature Cited
Kelley, D. (2015). Ocedata: Oceanographic datasets for oce. Retrieved from https://CRAN.R-project.org/package=ocedata
Kelley, D., & Richards, C. (2018). Oce: Analysis of oceanographic data. Retrieved from https://CRAN.R-project.org/package=oce
R Core Team. (2018). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Retrieved from https://www.R-project.org/