Introduction: A Brief Overview
The amount of data being generated by satellites has soared in recent years. The proliferation of remote sensing data can be explained by recent advancements in satellite technologies. However, according to Febvre1, this advancement set another challenge of handling and processing pentabyte of data satellite generates. Thanks to ERDDAP, for being in front-line to overcome this challenge. We can obtain satellite data within the geographical extent of the area we are interested. The ERDDAP server provides simple and consistent way to subset and download oceanographic datasets from satellites and buoys to your area of interest. ERDDAP is providing free public access to huge amounts of environmental datasets. Current the ERDDAP server has a list of 1385 datasets.This server allows scientist to request data of a specific area of interest in two forms—grids or tabular data.
xtractomatic is R package developed by Roy Mendelssohn2 that works with ERDDAP servers. The xtractomatic functions were originally developed for the marine science, to match up ship track and tagged animals with satellites data. Some of the satellite data includes sea-surface temperature, sea-surface chlorophyll, sea-surface height, sea-surface salinity, and wind vector. However, the package has been expanded and it can now handle gridded environmental satellite data from ERDAPP server.
In this post I will show two main routine operations extractomatic package can do. The first one is to match up drifter observation data with modis sea surface temperature satellite data using xtracto function. The second operation involves xtracto_3D to extract gridded data of sea surface temperature and chlorophyll a, both acquired by MODIS sensors.
Packages Required for the Process
Besides xtractomatic package, I am going to load other packages for data procesing and visualization. These packages include tidyverse3, lubridate4, spData5, sf6, oce7, and insol8 that I will use to manipulate, tidy, analyse and visulize the extracted SST and chl-a data from MODIS satellite. The chunk below shows the package needed to accomplish the task in this post
## load the package needed for the routine
# require(DT)
require(tidyverse)
require(lubridate)
require(spData)
require(sf)
require(oce)
require(insol)
require(xtractomatic)
# require(pals)Area of Interest: Pemba Channel
I chose the Pemba channel figure 1 to demonstrate the process of obtaining the environmental and oceanographical data from ERDDAP server. Briefly, the Pemba channel is the deepest channel when compared to other two channel found in Tanzania water—Zanzibar and Mafia channel. Because of deep bottom topography, the water in the Pemba channel is the least and oceanographic information in this channel is a formidable hurdle. I decided to use free satellite data and open-source sofware to bring up the environmental dynamics in this important channel. The channel is known for its small pelagic fishery that support local people as source of food and income.
## Map the Pemba channel with ggplot2, sf and ggsn packages
ggplot()+geom_sf(data = tz.ke, fill = "ivory", col = 1)+
coord_sf(xlim = c(38.5, 40), ylim = c(-6,-4))+
theme_bw()+
theme(panel.background = element_rect(colour = 1, fill = "lightblue"),
panel.grid = element_line(colour = NA),
axis.text = element_text(colour = 1, size = 10))+
scale_x_continuous(breaks = seq(38.5, 40, length.out = 4)%>%round(digits = 1))+
scale_y_continuous(breaks = seq(-5.8, -4.1,length.out = 5)%>%round(digits = 1))+
labs(x = NULL, y = NULL)+
geom_text(aes(x = 39.36, y = -5.2, label = "Pemba\nChannel"), col = "black")+
ggsn::scalebar(location = "bottomright", x.min = 38.5,
x.max = 39.95, y.min = -6, y.max = -4, dist = 25, dd2km = T,
model = "WGS84",st.dist = 0.02, st.size = 4)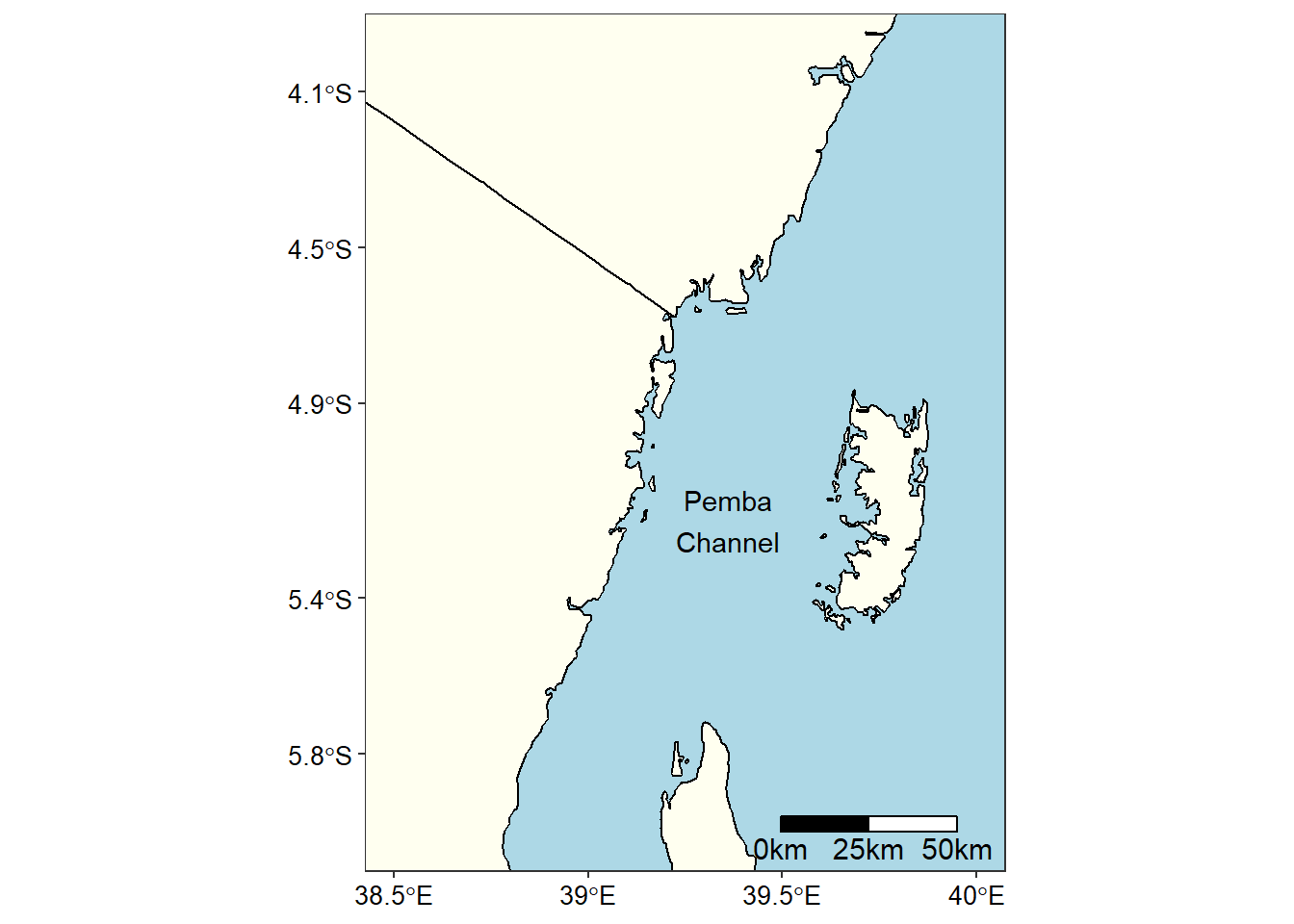
Figure 1: A map showing the location of the Pemba channel in the Indian Ocean Region (WIO)
Argo Floats serve as tagging features
Argo data provide sufficient information for tracking the water masses and the physical propoerties of the upper layer of the world oceans. Argo float with World Meteorological Identification number 1901124 that crossed the coastal water of Tanzania was downloaded as the actual Argo NetCDF files from the Argo Data website. Processing Argo float data I used oce package developed by Dan Kelley and Clark Richards. The oce package has read.argo function that read directly NetCDF file from Argo floats.
## read argo file
argo.1901124 = read.argo("E:/Doctoral/udsm/Processing/argo_profile/csiro/1901124/1901124_prof.nc")%>%handleFlags()
## make a section using the profiles recorded in argo float
argo.section = argo.1901124%>%as.section()Once the argo float profile data was imported in the workspace, was converted to hydrographic section that was used to plot sections of temperature against time (Figure 2) and temperature against longitude (Figure 3) from the surface to 1000 meter deep.
argo.section%>%plot(which = "temperature", ztype = "image", xtype = "time", ylim = c(1000,0))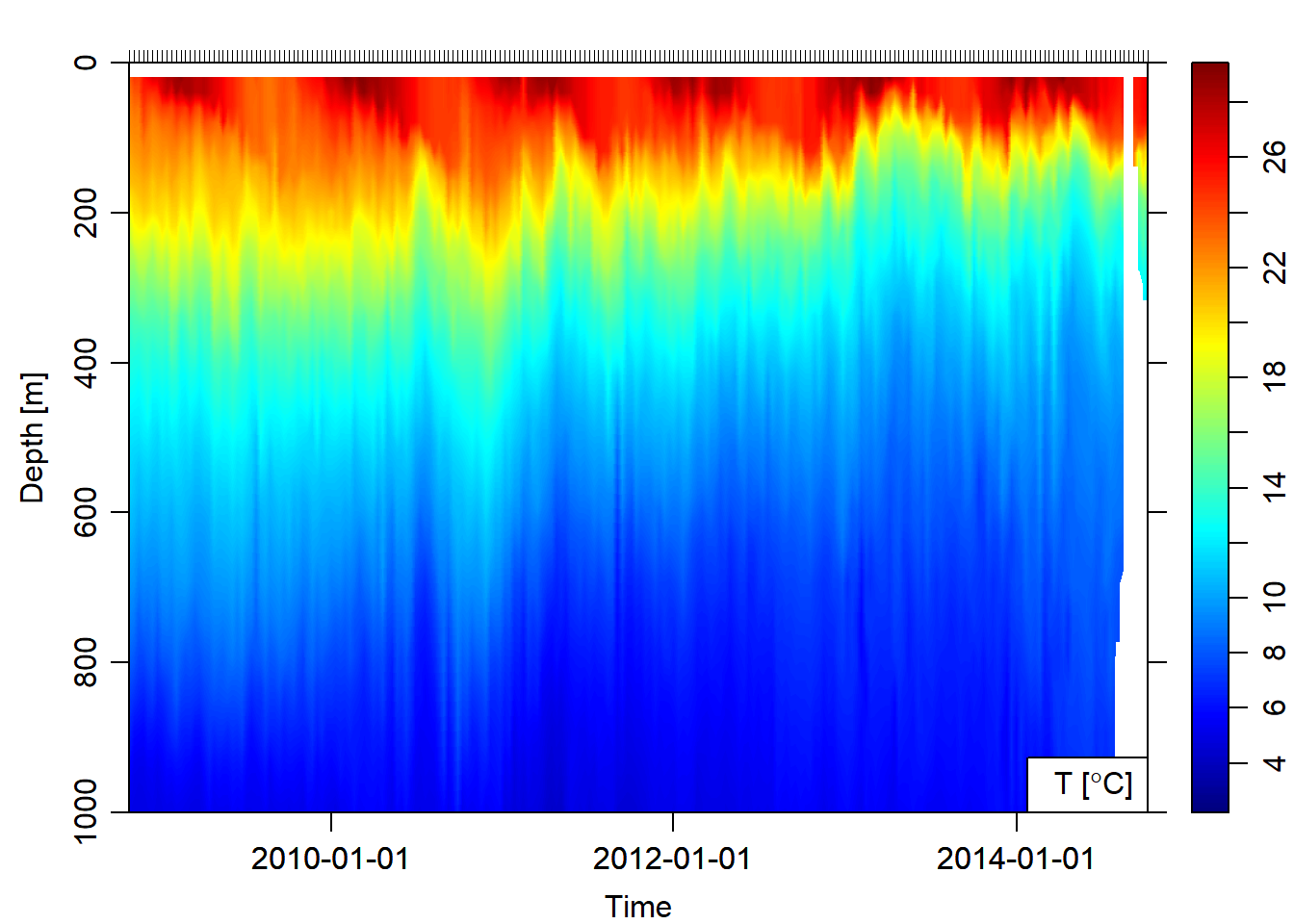
Figure 2: Hydrographic Section of Temperature obtained after combining float profiles of Argo float
argo.section%>%plot(which = "temperature", ztype = "image", xtype = "longitude", ylim = c(1000,0))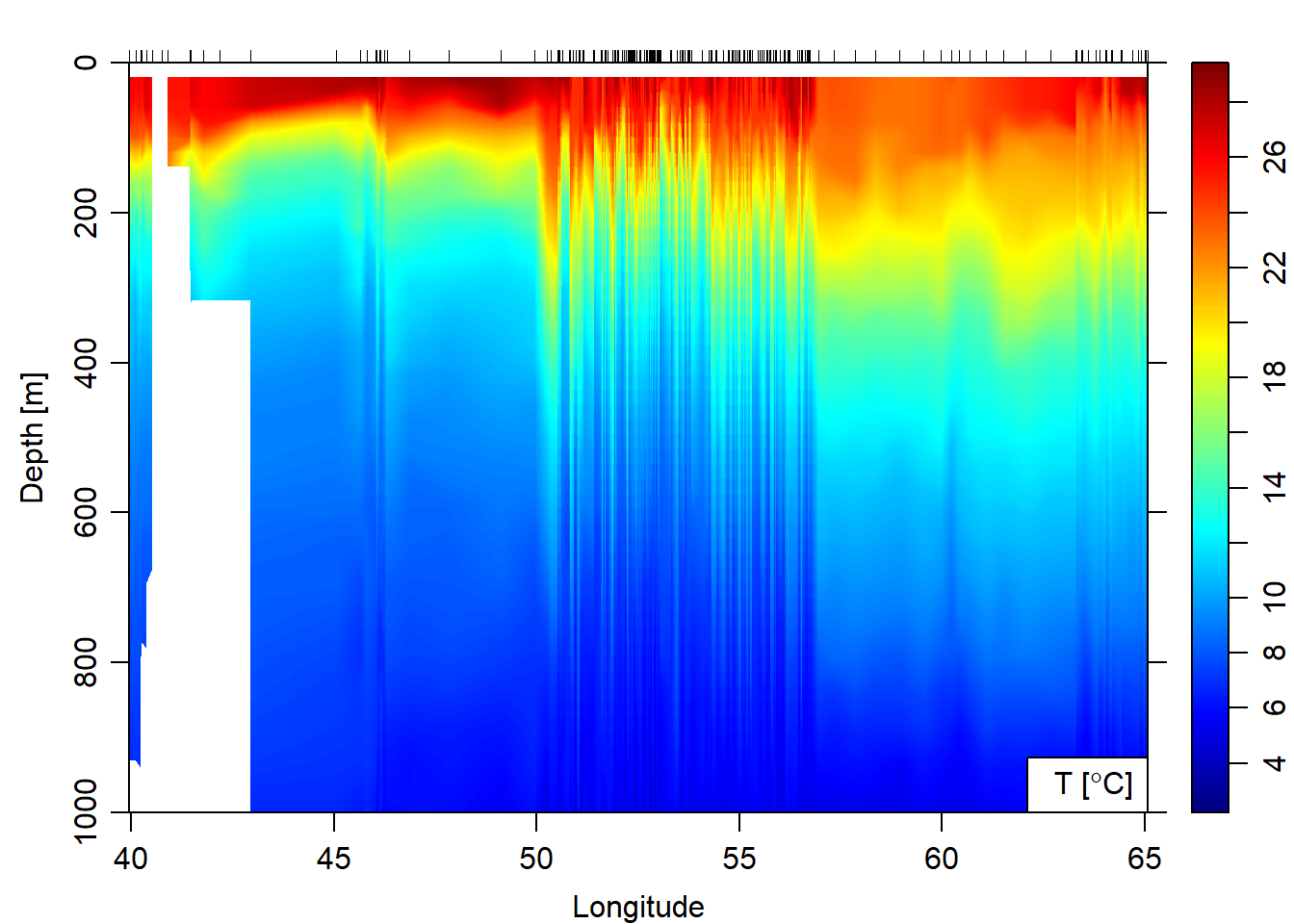
Figure 3: Hydrographic Section of Salinity obtained after combining float profiles of Argo float
To obtain Argo floats profile within the Pemba channel, I subset based on location and drop all profiles that were outside the geographical extent of the channel. Figure 4 show the six stations that the argo floats made vertical measurements of temperature and salinity close to the Pemba channel during the southeast monsoon season.
par(mfrow = c(1,2))
argo.section%>%subset(latitude<=-3.5 & longitude < 41)%>%plot(ztype = "contour", xtype = "time", ylim = c(500,0), showStations = T, which = c("map","temperature"), clongitude = c(35, 41))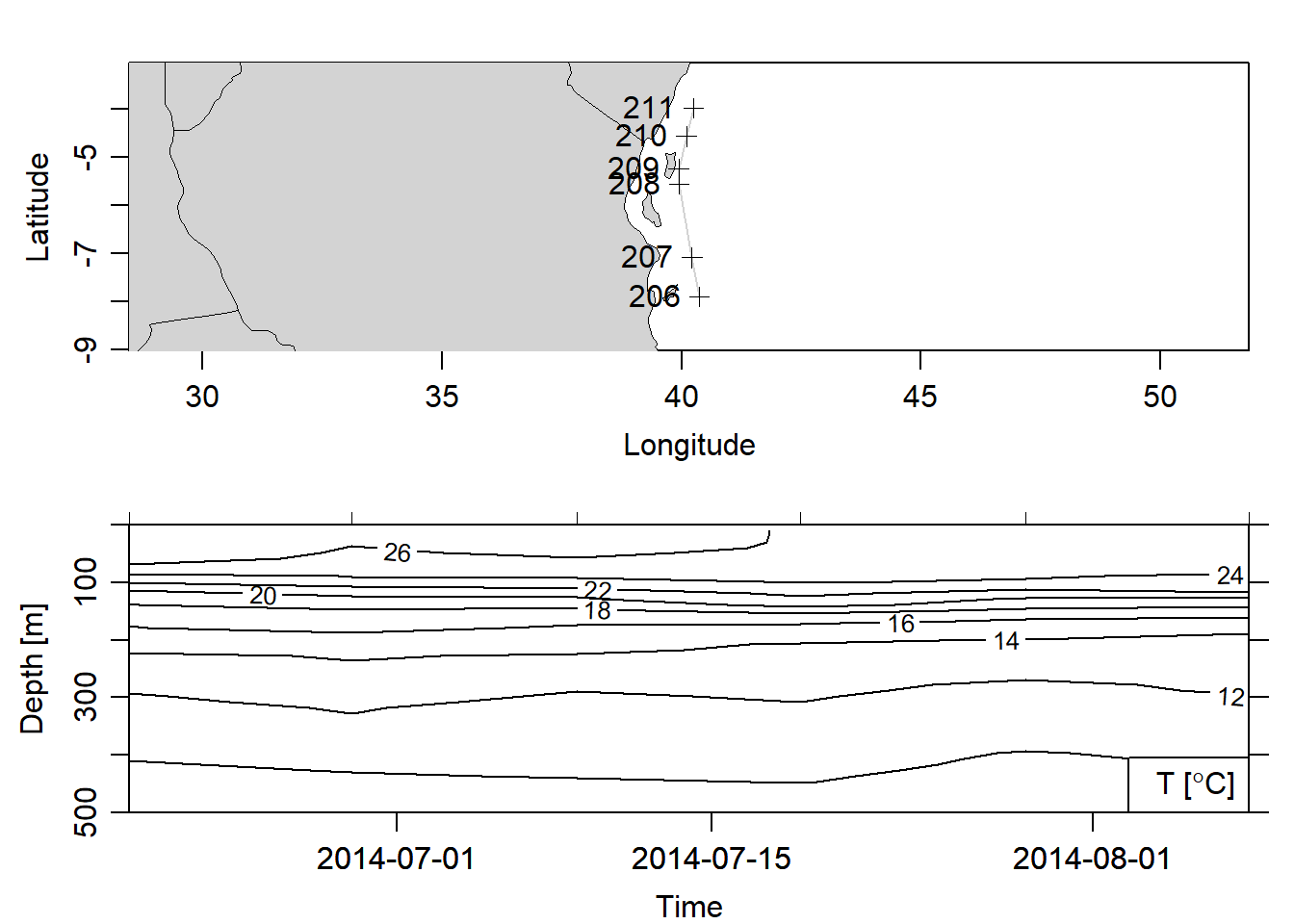
Figure 4: Section at Pemba channel
Make A tibble from Argo Section
A tibble is a modern data frame. Hadley Wickham declared that to change an old language like R and its traditional data fame that were useful 10 or 20 years ago, depend on the innovations in packages. One of those packages is the ecosystem of tidyverse that use pipe(%>%) operator and tibble for its operations. To make a tibble out of argo section. I converted the section into a list and then extracted the longitude and latitude information. I found that time is stored in the metadata slot and not the data slot. Being in different slot make it hard to extract
## convert argo section to list
argo.list = argo.section[["station"]]
## extract lon from the argo list
longitude = argo.section[["longitude", "byStation"]]
## extract lat from the argo list
latitude = argo.section[["latitude", "byStation"]]
## time can not be extracte the same way lon and lat can. This is because the time is stored in the
time = argo.section[["time", "byStation"]]Once I have the argo list fromt the section dataset, I plot the profile of the five casts measured close to the Pemba channel (Figure 5). These cast are labelled with station number 206 to 2011 from 2014-06-19 to 2014-08-07
par(mfrow = c(2,3))
argo.list[[206]]%>%plot(which = 1)
argo.list[[207]]%>%plot(which = 1)
argo.list[[208]]%>%plot(which = 1)
argo.list[[209]]%>%plot(which = 1)
argo.list[[210]]%>%plot(which = 1)
argo.list[[211]]%>%plot(which = 1)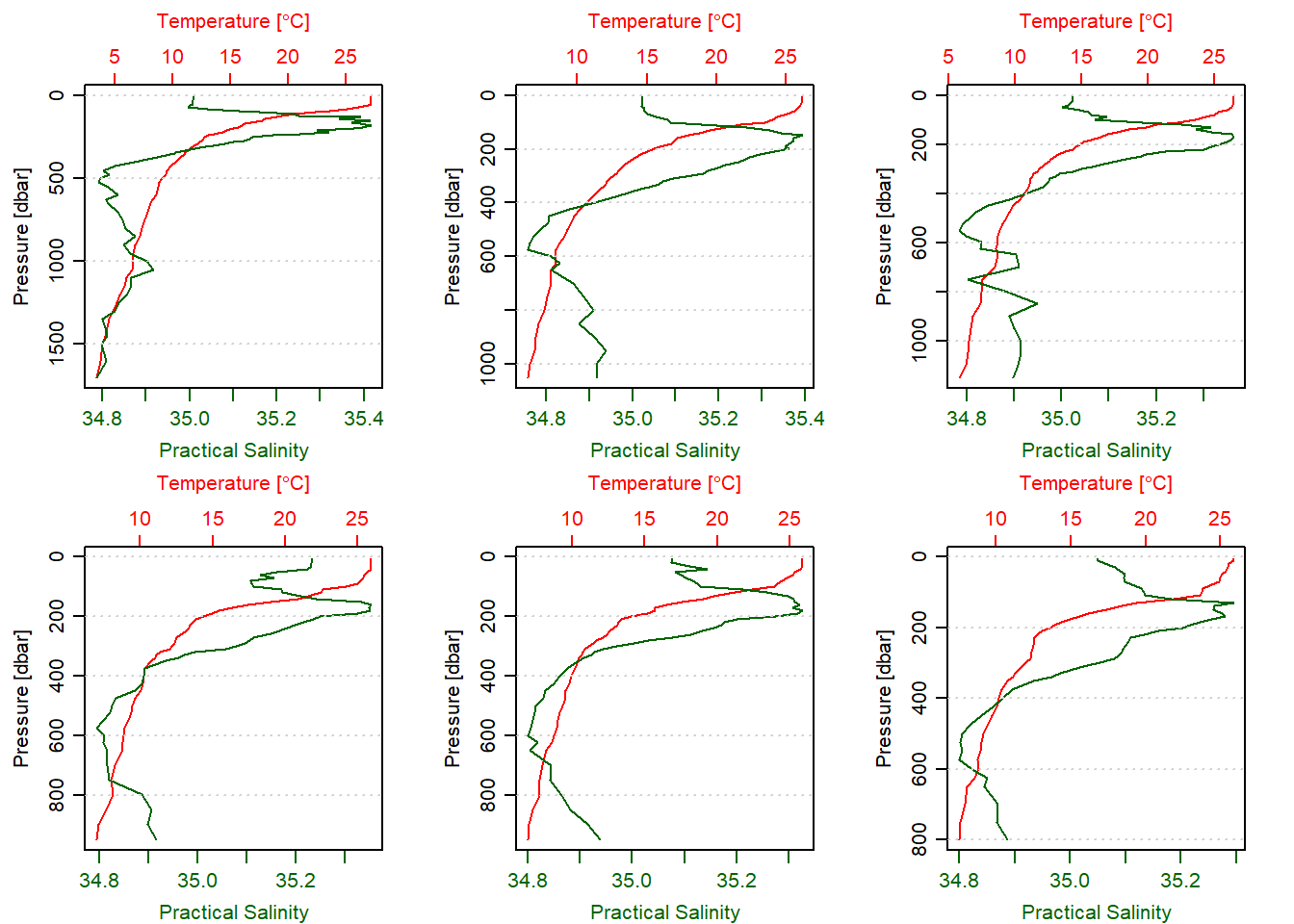
Figure 5: Temperature profile off the Pemba Island measured wth Argo float between
## extract Argo float profiles of station 206
profile.01 = argo.list[[206]]@data%>%
as.data.frame()%>%
as.tibble()%>%
mutate(Date = argo.list[[206]]@metadata$startTime%>%as_datetime(tz = ""),
Longitude = argo.list[[206]]@metadata$longitude,
Latitude = argo.list[[206]]@metadata$latitude)%>%
separate(Date, c("Date", "Time"),sep = " ", remove = TRUE)%>%
dplyr::select(Scan = scan, Date, Time, Longitude, Latitude,
Depth = pressure, Temperature = temperature,
Salinity = salinity)Table 1 shows the temperature and salinity from the surface to 800 meter deep of station 206 at this location 7.909S 40.372E during the southeast monsoon period of 2014-06-19. Although this cast contain 71 scans only the first ten rows are shown.
knitr::kable(profile.01%>%slice(1:10), caption = "Table 1: Overview of CTD measurement of first cast of Argo float number 1901124", digits = 2, align = "c", row.names = FALSE, escape = T)| Scan | Date | Time | Longitude | Latitude | Depth | Temperature | Salinity |
|---|---|---|---|---|---|---|---|
| 1 | 2014-06-19 | 04:30:45 | 40.37 | -7.91 | 4.0 | 27.16 | 35.01 |
| 2 | 2014-06-19 | 04:30:45 | 40.37 | -7.91 | 9.4 | 27.16 | 35.01 |
| 3 | 2014-06-19 | 04:30:45 | 40.37 | -7.91 | 19.2 | 27.16 | 35.01 |
| 4 | 2014-06-19 | 04:30:45 | 40.37 | -7.91 | 28.8 | 27.17 | 35.01 |
| 5 | 2014-06-19 | 04:30:45 | 40.37 | -7.91 | 39.5 | 27.17 | 35.01 |
| 6 | 2014-06-19 | 04:30:45 | 40.37 | -7.91 | 49.6 | 27.15 | 35.01 |
| 7 | 2014-06-19 | 04:30:45 | 40.37 | -7.91 | 59.6 | 26.92 | 35.00 |
| 8 | 2014-06-19 | 04:30:45 | 40.37 | -7.91 | 69.6 | 26.02 | 35.00 |
| 9 | 2014-06-19 | 04:30:45 | 40.37 | -7.91 | 79.3 | 25.04 | 35.04 |
| 10 | 2014-06-19 | 04:30:45 | 40.37 | -7.91 | 89.5 | 23.58 | 35.11 |
I extracted the argo float information for station 206 and there are 217 casts made by this float. Manual extraction of CTD information one cast after the other is tedious and time consuming. Because of the danger of manual extraction of CTD information from argo float, I use a For loop to iterate the process for all 217 casts.
argo.tb = NULL
for (i in 1:217){
profile = argo.list[[i]]@data%>%
as.data.frame()%>%
as.tibble()%>%
mutate(Date = argo.list[[i]]@metadata$startTime%>%as_datetime(tz = ""),
Longitude = argo.list[[i]]@metadata$longitude,
Latitude = argo.list[[i]]@metadata$latitude)%>%
separate(Date, c("Date", "Time"),sep = " ", remove = TRUE)%>%
dplyr::select(Scan = scan, Date, Time, Longitude, Latitude,
Depth = pressure, Temperature = temperature,
Salinity = salinity)
argo.tb = argo.tb%>%bind_rows(profile)
}Now I have a tibble of 15407 scan from 2017 measured from the surface to the deepest part at a particular location. However, I want only the surface information of salinity, and temperature from each cast. I used filter function from dplyr9 package to retain the first scan in each cast, which are measured at the surface (Table 2) and its trajectory figure 6
argo.surface.tb = argo.tb%>%filter(Scan == 1)
knitr::kable(argo.surface.tb%>%slice(1:10), caption = "Table 2: Surface Argo floats measurements from Argo float number 1901124", digits = 2, align = "c", row.names = FALSE, escape = T)| Scan | Date | Time | Longitude | Latitude | Depth | Temperature | Salinity |
|---|---|---|---|---|---|---|---|
| 1 | 2008-10-28 | 01:43:57 | 64.03 | -21.64 | 4.6 | 23.55 | 35.34 |
| 1 | 2008-11-07 | 02:17:07 | 64.20 | -21.28 | 4.3 | 24.59 | 35.04 |
| 1 | 2008-11-17 | 02:30:18 | 64.07 | -21.01 | 4.4 | 24.99 | 35.02 |
| 1 | 2008-11-27 | 02:37:35 | 63.61 | -21.10 | 4.7 | 25.71 | 34.98 |
| 1 | 2008-12-07 | 05:03:59 | 63.43 | -21.25 | 4.4 | 26.10 | 35.03 |
| 1 | 2008-12-17 | 03:03:40 | 63.32 | -21.46 | 4.8 | 27.04 | 35.13 |
| 1 | 2008-12-27 | 05:09:53 | 63.47 | -21.56 | 4.6 | 26.72 | 35.08 |
| 1 | 2009-01-06 | 02:40:37 | 63.90 | -21.60 | 4.6 | 28.40 | 35.08 |
| 1 | 2009-01-16 | 04:57:04 | 64.45 | -21.49 | 4.7 | 27.99 | 35.12 |
| 1 | 2009-01-26 | 02:30:53 | 64.83 | -21.21 | 4.6 | 28.07 | 34.96 |
data("world") ## medium resolution from spData package
ggplot()+
geom_sf(data = world, fill = "ivory", col = 1)+
geom_path(data = argo.surface.tb,
aes(x = Longitude, y = Latitude), size =.5, col = 2)+
geom_point(data = argo.surface.tb, aes(x = Longitude, y = Latitude), col = 2)+
coord_sf(xlim = c(35, 71), ylim = c(-25,0))+
ggrepel::geom_text_repel(data = argo.surface.tb%>%slice(seq(3,217,25)),
aes(x = Longitude, y = Latitude, label = Date))+
theme_bw()+
theme(panel.background = element_rect(colour = 1, fill = "lightblue"),
panel.grid = element_line(colour = NA),
axis.text = element_text(colour = 1, size = 10))+
labs(x = NULL, y = NULL)+
ggsn::scalebar(location = "bottomright", x.min = 35,
x.max = 70, y.min = -25, y.max = 0, dist = 500, dd2km = T,
model = "WGS84",st.dist = 0.02, st.size = 4)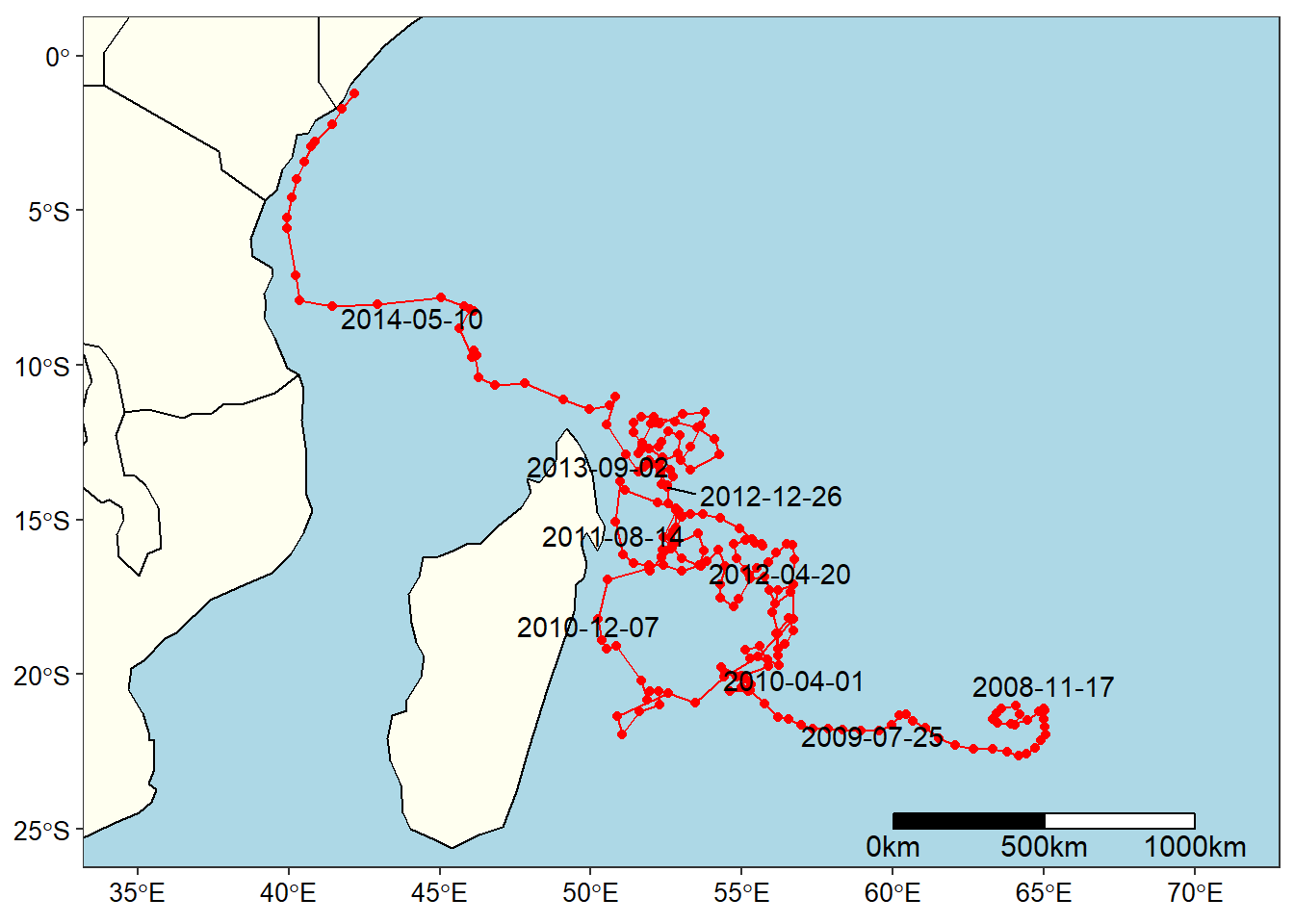
Figure 6: TRajectory of Argo float ID 1901124
Tracking Satellite Data with Argo Float as Tagged Data
The argo.surface.tb dataset consist of eight variables—scan, date, time, longitude, latitude, depth, temperature and salinity of 216 casts from Argo float with ID 1901124 measured from 2008-10-28 to 2014-09-27 in the Indian Ocean. The date, longitude, and latitude variables from this dataset was used as xpos, ypos and tpos arguments in the xtracto function.
require(xtractomatic) ## load the package
modis.sst = xtracto(dtype = "mhsstd8day",
xpos = argo.surface.tb$Longitude,
ypos = argo.surface.tb$Latitude,
tpos = argo.surface.tb$Date, ,#note a space here.A bug!!!!
xlen = 0.2,
ylen = 0.2)
write_csv(modis.sst, "modis_argo_match.csv")Tidying the data
Once the data has been downloaded was saved in my working directory and parsed a command that prevent the execution of the chunk whenever we run the script. This dataset was then loaded as tibble in the workspace in the workspace. Because the dimension of the argo and modis tibble files are the same, it was easy to bind them together and then select some variables of interest of further and analysis (Table 3).
## load the dataset
modis.sst = read_csv("E:/Data Manipulation/xtractomatic/modis_argo_match.csv")%>%
mutate(Date = as.Date(`satellite date`))
## make character date in argo.surface.tab as date format
argo.surface.tb$Date = as.Date(argo.surface.tb$Date)
## tidy the data
modis.argo.tb = argo.surface.tb%>%
dplyr::select(-c(Scan,Time, Salinity))%>%
bind_cols(modis.sst%>%
dplyr::select(satelliteDate = Date, n,
satelliteMeanSST = `mean sst`))%>%
dplyr::select(Date, Longitude, Latitude, Temperature ,satelliteDate, n, satelliteMeanSST)
knitr::kable(modis.argo.tb%>%sample_n(20), caption = "Table 3: Tidied drifter and modis temperature data", digits = 2, align = "l")# write_csv(modis.argo.tb, "modis_argo_tb.csv")
modis.argo.tb = read_csv("E:/Data Manipulation/xtractomatic/modis_argo_tb.csv")## Parsed with column specification:
## cols(
## Date = col_date(format = ""),
## Longitude = col_double(),
## Latitude = col_double(),
## Temperature = col_double(),
## satelliteDate = col_date(format = ""),
## n = col_integer(),
## satelliteMeanSST = col_double()
## )knitr::kable(modis.argo.tb%>%sample_n(20), caption = "Table 3: Tidied drifter and modis temperature data", digits = 2, align = "l")| Date | Longitude | Latitude | Temperature | satelliteDate | n | satelliteMeanSST |
|---|---|---|---|---|---|---|
| 2014-01-20 | 49.11 | -11.10 | 28.70 | 2014-01-21 | 7 | 28.40 |
| 2009-06-15 | 61.52 | -22.08 | 24.65 | 2009-06-14 | 29 | 24.59 |
| 2011-10-13 | 52.81 | -15.25 | 24.90 | 2011-10-12 | 33 | 24.44 |
| 2012-05-20 | 55.50 | -16.58 | 27.09 | 2012-05-20 | 36 | 26.99 |
| 2011-05-06 | 52.39 | -15.58 | 28.32 | 2011-05-05 | 30 | 28.02 |
| 2013-07-14 | 52.79 | -11.84 | 25.53 | 2013-07-16 | 34 | 25.55 |
| 2009-11-12 | 55.28 | -20.52 | 25.55 | 2009-11-13 | 30 | 25.48 |
| 2009-01-26 | 64.83 | -21.21 | 28.07 | 2009-01-29 | 36 | 28.58 |
| 2009-11-02 | 55.77 | -20.94 | 25.13 | 2009-11-05 | 24 | 25.72 |
| 2010-09-18 | 50.89 | -21.36 | 24.47 | 2010-09-18 | 25 | 24.67 |
| 2010-12-27 | 50.35 | -18.89 | 28.26 | 2010-12-29 | 30 | 28.48 |
| 2013-08-03 | 51.67 | -11.68 | 25.04 | 2013-08-01 | 20 | 24.63 |
| 2011-11-12 | 53.00 | -16.25 | 26.22 | 2011-11-13 | 36 | 26.04 |
| 2013-06-04 | 53.31 | -13.38 | 26.31 | 2013-06-06 | 25 | 25.42 |
| 2012-10-17 | 52.56 | -14.48 | 25.47 | 2012-10-19 | 30 | 24.24 |
| 2014-01-30 | 47.83 | -10.58 | 28.81 | 2014-01-29 | 36 | 29.05 |
| 2014-06-29 | 40.22 | -7.10 | 26.21 | 2014-06-30 | 36 | 26.21 |
| 2009-03-27 | 64.91 | -22.14 | 27.64 | 2009-03-26 | 30 | 27.67 |
| 2014-04-30 | 45.81 | -8.11 | 28.20 | 2014-04-27 | 30 | 27.86 |
| 2011-02-15 | 52.35 | -16.20 | 25.03 | 2011-02-14 | 10 | 24.80 |
rsq = cor(modis.argo.tb$Temperature, modis.argo.tb$satelliteMeanSST)%>%
round(2)
fit = lm(modis.argo.tb$Temperature~modis.argo.tb$satelliteMeanSST, method = "qr", model = TRUE)
summary(fit)##
## Call:
## lm(formula = modis.argo.tb$Temperature ~ modis.argo.tb$satelliteMeanSST,
## method = "qr", model = TRUE)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.22086 -0.20670 -0.02381 0.22081 2.04826
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.86593 0.45913 4.064 6.77e-05 ***
## modis.argo.tb$satelliteMeanSST 0.93127 0.01718 54.194 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.4388 on 214 degrees of freedom
## Multiple R-squared: 0.9321, Adjusted R-squared: 0.9318
## F-statistic: 2937 on 1 and 214 DF, p-value: < 2.2e-16Accuracy Measures
accuracy = pracma::rmserr(x = modis.argo.tb$Temperature,
y = modis.argo.tb$satelliteMeanSST,
summary = FALSE)To understand the accuracy of surface temperature measured with argo floats and satellite, I used a prominent root mean square error (RMSE) technique. A pracma package has rmserr function that deals with accuracy assessment of six different types. The result showed the two data collection technique are fairly very close with an accuracy of 0.454053 degree centigrade
The correlation coefficient of surface temperature measured by Argo and MODIS was found to be R^2 = 0.9321 (Figure 7).This correlation indicate that the two data sources have significant association of about 93 percent (p < 0.05). Therefore, we can trust the sea surface temperature data from MODIS sensor as its measurements are close to those collected in-situ with Argo floats.
ggplot(data = modis.argo.tb,
aes(x = Temperature, y = satelliteMeanSST))+
geom_point()+
geom_smooth(method = "lm", col = 2, se = FALSE)+
theme(panel.background = element_rect(colour = 1),
axis.text = element_text(colour = 1, size = 12),
axis.title = element_text(colour = 1, size = 14))+
geom_text(aes(x = 29, y = 24,
label = paste("RSq = ", 0.93)), size = 4)+
scale_x_continuous(breaks = seq(23.5,29.5, 1.5))+
scale_y_continuous(breaks = seq(23,31, 1.5))+
labs(y=expression(~Modis~Sensor~SST~(~degree~C)), x=expression(~Argo~float~Temperature~(~degree~C)))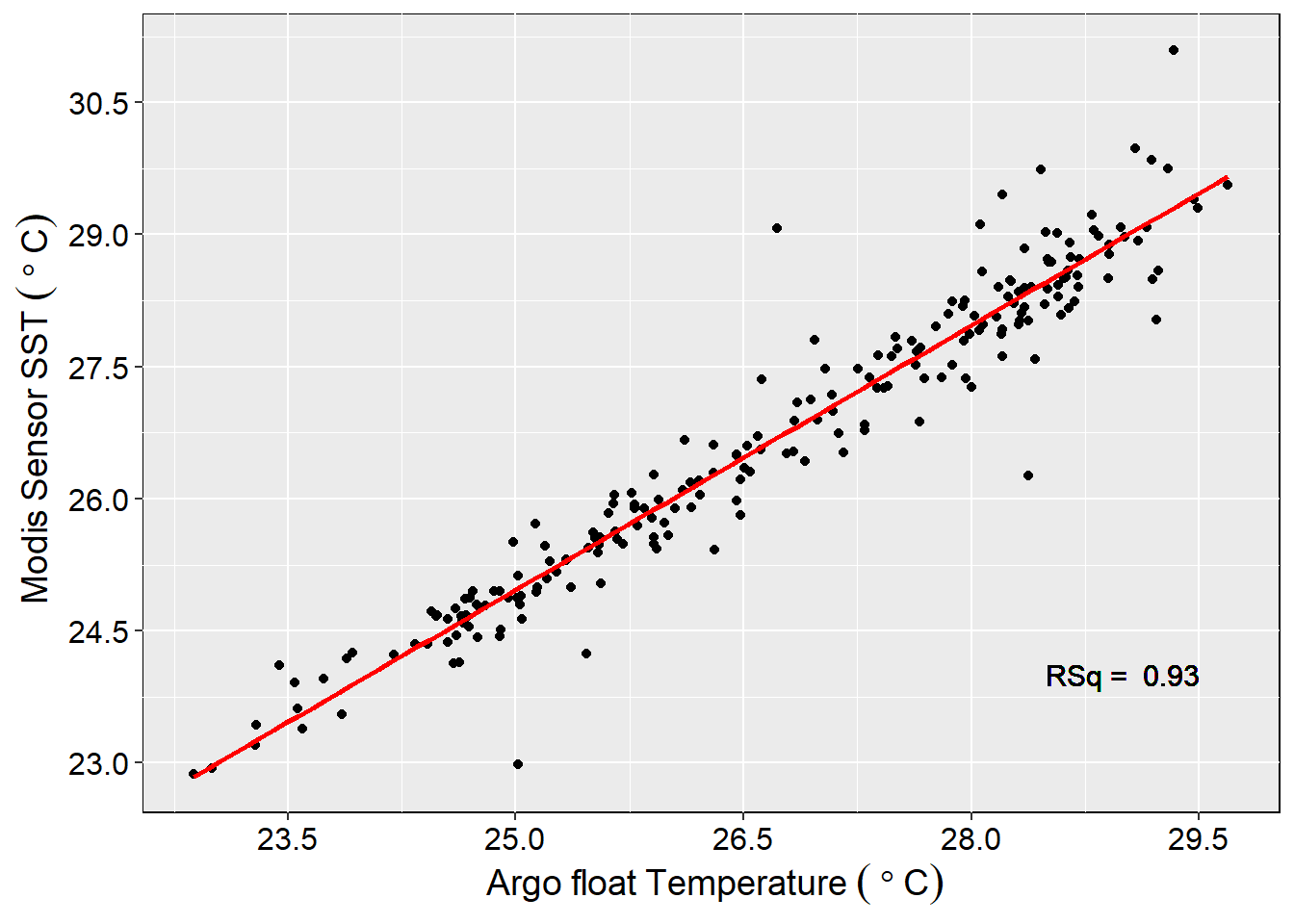
Figure 7: Correlation of temperature between Argo float and MODIS surface temperature
Cited Articles
Paul Febvre, CTO of the Satellite Applications Catapult (SAC), which aims to bring together academia and businesses, as well as opening the sector to new markets↩
Mendelssohn, R. (2017). xtractomatic: accessing environmental data from ERD’s ERDDAP server. R package version, 3(2)↩
Wickham, H. (2017). Tidyverse: Easily install and load’tidyverse’packages. R package version, 1(1)↩
Grolemund, G., & Wickham, H. (2013). lubridate: Make dealing with dates a little easier. R package version, 1(3).↩
Lovelace, R., Nowosad, J., & Muenchow, J. Geocomputation with R.↩
Pebesma, E. (2016). sf: Simple Features for R.↩
Kelley, D., & Richards, C. (2017). oce: Analysis of Oceanographic Data. R package version 0.9-22.↩
Corripio, J. G. (2014). Insol: solar radiation. R package version, 1(1), 2014.↩
Wickham, H., Francois, R., Henry, L., & Müller, K. (2018). dplyr: A grammar of data manipulation. R package version 0.7, 6.↩